Adduct nomenclature
Adduct information can be provided in two ways:
- Specified in the input file created by third-party preprocessing tools (using peak list-based formats such as
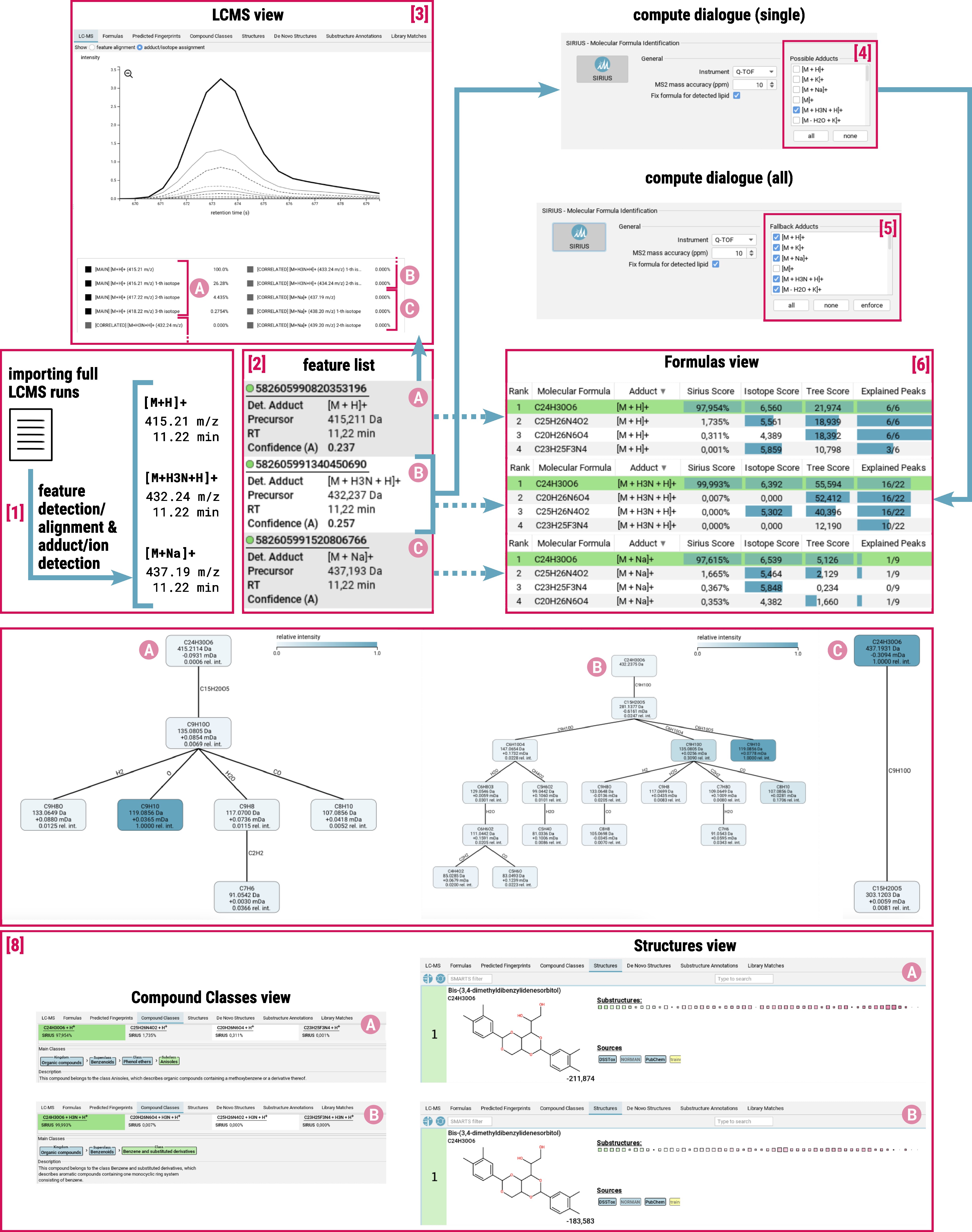
.mgf). - SIRIUS can import full LCMS runs in
.mzMLand.mzXMLformats through its pre-processing tool. It performs feature detection and alignment based on MS/MS spectra, as well as adduct detection and assigns ion types with specific adducts or in-source losses to the features [1].
The specified adduct will affect the possible molecular formula candidates [6] of a feature and consequently all subsequent computations [8] (fingerprint prediction, compound class prediction, molecular structure hit) .
SIRIUS uses the following nomenclature to designate losses, adducts, and ionizations:
[M-LOSS+ADDUCT±IONIZATION]±
Specific cases:
- Positive ions:
[M+IONIZATION]+, such as[M+H]+,[M+Na]+ - Negative ions:
[M±IONIZATION]−, such as[M-H]-,[M+Cl]- - Intrinsically charged compounds:
[M]+or[M]- - Unknown losses or adducts:
[M+?]+or[M+?]-for positive / negative ions with unspecified modifications - Complex cases:
[M - H + Na + Na]+indicates a hydrogen loss (-H), a sodium adduct (+Na), and ionization by sodium (+Na+)
The most common ionization modes are [M+H]+, [M+Na]+, [M-H]-, [M+Cl]-.
Import:
Currently, SIRIUS supports only singly charged compounds. Hence, adducts such as [M+2H]2+ will be skipped during import. Additionally, dimers, which are represented by [2M], are also not yet supported and will be skipped during import. Adducts will be parsed to fit our nomenclature, e.g. [M+H4N]+ will be parsed to [M+H3N+H]+.
Detecting addutcs in LCMS pre-processing
Pre-processing [1] detects adducts from a predefined list and selects them based on correlating chromatographic peaks with indicative mass differences in the data. Detected features are listed in the feature list [2] on the left side of the GUI. A feature contains all MS and MS/MS spectra corresponding to a measured aligned feature. For each feature, the detected adduct type(s) are shown in this panel.
It is often challenging to unambiguously assign a single adduct to every feature.
- In case a single adduct is assigned to the feature, this adduct is displayed in the feature list and will be used for computations.
- In case the adduct assignment is ambiguous, all possible adducts are listed in the feature list and SIRIUS will consider multiple adducts for computations.
- In case no adduct could be assigned, the feature list displays
[M+?]+. For computations, a set of fallback adducts is used which can be specified by the user.
The feature list can be filtered for features with specific adducts. Use th filter button (three dots to the right of the search field) to open the filter dialog. In the Input tab, select the adducts you would like to filter for.
The LC-MS view [3] displays the ion chromatogram of a detected feature. You can choose whether to show the feature alignment or the adduct/isotope assignment. In the adduct/isotope assignment view, you can find the merged mass trace of the aligned feature (bold black line) alongside its isotopes (dashed black lines) (A) and correlated features with different adducts (grey lines) alongside their isotopes (dashed grey lines) (B) and (C).
Molecular formula annotation
To identify molecular formulas, you can specify the adducts to be considered. Be aware that there is a difference in the compute dialog for doing computations on a single feature or on all features.
- Single-feature compute dialog: Select all adducts that will be considered for computation in the
Possible Adductsfield [4]. The detected adducts are pre-selected. In case no adduct could be assigned, SIRIUS default adducts are pre-selected. - All-features compute dialog: In the
Fallback Adductsfield [5] you can specify fallback adducts that will be used for all features for which no adducts were detected during preprocessing (or prior external annotation). Using the enforce option, you can even enforce to consider the selected adducts for all features (in addition to the detected adducts).
!! Refrain from selecting all adducts as fallback adducts !! Multiple testing inflates the chance of errors. Adding more fallback adducts to the pool increases the probability of producing a false positive. You should only select fallback adducts that you think might actually be present in the data. For example, if you know that a certain type of sample is prevalent to a certain adducts you can add it to the list of fallback adducts. You may manually try other adducts for single compounds using the single-feature compute dialog.
Be aware that formula annotation (and thus subsequent tools as well) will additionally consider the “base ionization”. For example, if the preprocessing assigns [M+H2O+K]+, formula annotation will also consider [M+K]+. However, SIRIUS only displays [M+H2O+K]+ as detected adduct for the input data, while [M+K]+ is only part of the results (formula annotation and so on).
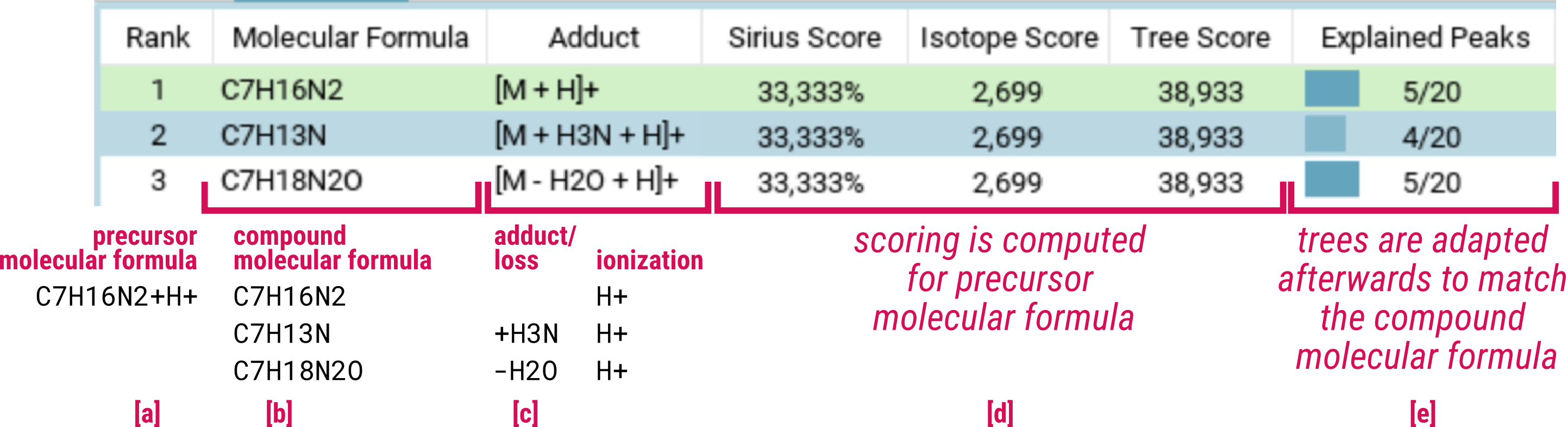
During the formula annotation step, SIRIUS generates and scores molecular formula candidates explaining the precursor mass. A precursor molecular formula [a] can be explained by different compound molecular formulas [b] using different adduct candidates [c].
All adducts of the same precursor formula receive identical scores [d], since it is not possible to distinguish between them based on the isotope pattern and MS/MS spectrum: The isotope patterns will be identical and a loss observed in the MS/MS spectrum could stem from either the adduct or a covalently bonded part of the molecule.
Two specific details must be noted:
- Fragmentation trees which are used to score molecular formula candidates, are provided in neutral form. For all adducts with the same ionization (e.g. [M+H]+, [M-H2O+H]+ and [M+H3N+H]+), a common fragmentation tree is computed. Then, fragmentation trees are resolved for each specific adduct [e]/[7]. During this process, some fragments may not be explained by a resolved formula and are removed from the tree:
- Resolving C7H16N2 for adduct/ion [M+H3N+H]+ is possible (C7H13N)
- Resolving C6H12O6 for adduct/ion [M+H3N+H]+ is not possible
Despite removing these fragments, we do not alter the score for the fragmentation tree, as the fragment could have had another possible explanation, and we do not want to penalize the candidate due to this post-processing.
- We do not differentiate between [M+H]+ and [M]+. In LC-MS experiments [M]+ is very uncommon. Moreover, for an unknown compound in an untargeted measurement it is challenging to determine if the compound was intrinsically charged or ionized later by the instrument. Therefore, SIRIUS considers the same neutral molecular formula for both adducts (as [M+H]+), but also searches for intrinsically charged molecular structures at the database search step. Per default [M+H]+ is considered, and [M]+ is merely treated as a special case of [M+H]+. [M]+ is used if directly specified in the input file or by the user.
Exporting molecular formula results with multiple adducts
The summary file formula_identifications.[tsv|csv|xlsx] contains the top-ranked molecular formula for each feature, based on the SIRIUS score or ZODIAC score, if available.
When multiple adduct candidates correspond to the same precursor molecular formula (e.g. [C20H14O6+H3N+H]+ and [C20H19NO7-H2O+H]+),
they will have the identical score.
In such cases, the top-ranked candidate in formula_identifications.[tsv|csv|xlsx] is resolved
to [C20H17NO6+H]+ only considering the ionization but ignoring adducts.
The file formula_identifications_adducts.[tsv|csv|xlsx] resolves the top-ranked precursor molecular formula
for all detected adducts (in this case
[C20H14O6+H3N+H]+ and [C20H19NO7-H2O+H]+).